After read the answers of some similar questions on this site, e.g.,
Generate Correlated Normal Random Variables
Generate correlated random numbers precisely
I wonder whether such approaches can assure the specific distributions of random variables generated.
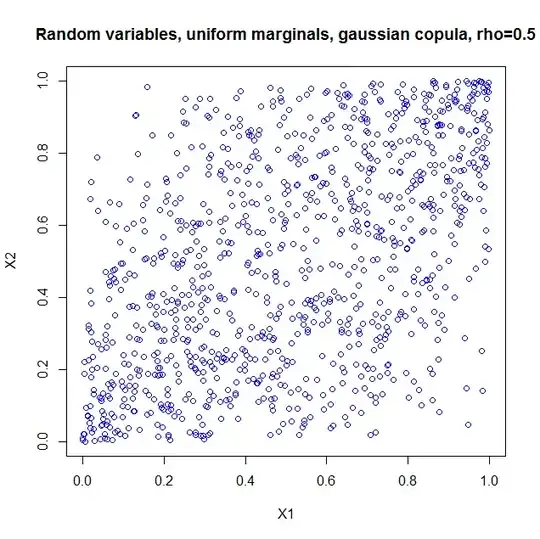
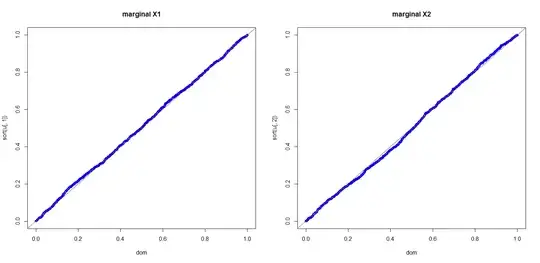
In order to make it easier to present my question, let us consider a simple case of creating correlated two uniform continuous random variables on $[0,1]$ with correlation coefficient $\dfrac{1}{2}=\rho$.
The methods by Cholesky decomposition (or spectral decomposition, similarly) first generates $X_1$ and $X_2$ which are independent pseudo random numbers uniformly distributed on $[0,1]$, and then creates $X_3=\rho X_1+\sqrt{1-\rho^2} X_2$. The $X_1$ and $X_3$ thus created are random variables with correlation coefficient $\rho$.
But the problem is, $X_3$ 's probability density fuction is triangle /trapezoid distribution which can be deducted by the convolution of the density functions of $X_1$ and $X_2$.
The probability density functions of $\rho X_1$ and $\sqrt{1-\rho^2} X_2$ are:
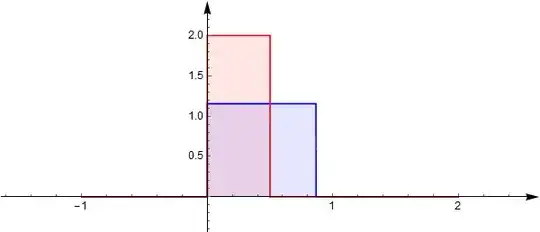
The convolution (sum) of them $X_3$ has density function:
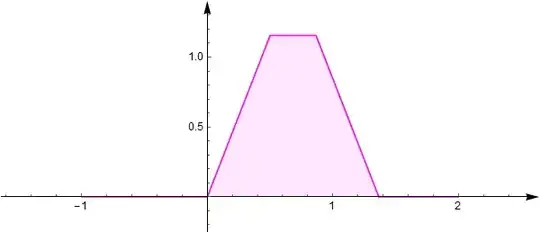
This means, the distribution of $X_3$ is not the desired uniform one on $[0,1]$.
What should I do in order to create random variables uniformly distributed on $[0,1]$ with correlation coefficient $\rho$ ?
The similar issue persists when I want to create multiple correlated random variables with predefined correlation matrix.
Considering the pseudo random variables usually are not really independent with a correlation coefficient between -1 and 1, it seems that: it is difficult to generate numerically independent $[0,1]$ uniform random variables since the uncorrelation transformation seems to always change the distribution profile.
PS: Before asking this question, I had read the following questions and links but didnot find an answer :
http://www.sitmo.com/article/generating-correlated-random-numbers/
http://numericalexpert.com/blog/correlated_random_variables/