I work with a Sentinel-2 .jp2 image (red band, 10950 x 10950 pixels). My aim is to reach the same result what SNAP does with a Python script. See the SNAP method and parameters:
So this is my reference (result with SNAP), I want to reach this result (QGIS grayscale representation, cumulative cut - 2/98%):
So I tried to replicate it with GDAL:
import numpy as np
from osgeo import gdal, gdal_array
input = "d:/bitbucket/cnn-lcm/T33TWM_A012703_20171127T100339_B04.jp2"
output = "d:/bitbucket/cnn-lcm/T33TWM_A012703_20171127T100339_B04_gdal.tif"
dataset = gdal.Open(input)
array = dataset.ReadAsArray()
percentile_025 = np.percentile(array, 2.5) # 349.0
percentile_975 = np.percentile(array, 97.5) # 3735.0
command = 'gdal_translate -scale ' + str(percentile_025) + ' ' + str(percentile_975)+ ' 0 255 -of GTiff -ot Byte' + ' ' + input + ' ' + output
os.system(command)
The GDAL result is not the same, its a bit brighter, the white areas are bigger. The values are not the same on the layers panel (QGIS grayscale representation, cumulative cut - 2/98%):
The Orfeo code:
import otbApplication
Convert = otbApplication.Registry.CreateApplication("Convert")
Convert.SetParameterString("in", "d:/bitbucket/cnn-lcm/T33TWM_A012703_20171127T100339_B04.jp2")
Convert.SetParameterString("out", "d:/bitbucket/cnn-lcm/T33TWM_A012703_20171127T100339_B04_hcut.tif")
Convert.SetParameterString("type","linear")
Convert.SetParameterString("hcp.high","2.5")
Convert.SetParameterString("hcp.low","2.5")
Convert.ExecuteAndWriteOutput()
Rescale = otbApplication.Registry.CreateApplication("Rescale")
Rescale.SetParameterString("in", "d:/bitbucket/cnn-lcm/T33TWM_A012703_20171127T100339_B04_hcut.tif")
Rescale.SetParameterString("out", "d:/bitbucket/cnn-lcm/T33TWM_A012703_20171127T100339_B04_orfeo.tif")
Rescale.SetParameterOutputImagePixelType("out", 1)
Rescale.SetParameterFloat("outmin", 0)
Rescale.SetParameterFloat("outmax", 255)
Rescale.ExecuteAndWriteOutput()
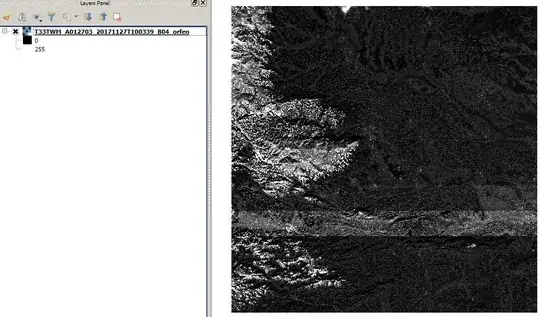
The Orfeo result is very similar to GDAL (only 1-2 value differences in pixels). And there are big, problematic strips in the middle (QGIS grayscale representation, cumulative cut - 2/98%):
So finally my questions:
Is it possible to eliminate the differences? Is it possible to reach exactly the result of SNAP? And how?
Download link to data: http://sentinel-s2-l1c.s3.amazonaws.com/tiles/33/T/WM/2017/11/27/0/B04.jp2



@ user1269942: It's a good idea, I'll test it as well, and modify the code.
– pnz1337 Nov 24 '18 at 10:51