I need to know the "number of clusters" in a dataaset.
To find the number of clusters, I am using a Gaussian Mixture model fitting,
bear with me,
Because the underlying distributions (each cluster) are not Gaussian, the GM tends to give very bad fits, because it is trying to compensate for the skewness of the data by increasing the variance of the Gaussian it fits and things like this. I figured, I may be able to solve this by fitting more Gaussians than are clusters expected to be in the data and then based on the distance of clusters find out which ones are true clusters and which ones are fits to the same cluster.
Now my problem is I have a distance matrix (Mahalanobis distances), distances between the fitted Gaussians coming out of the Gaussian mixture model, but I have no reliable way of counting the clusters,
to make it a bit more clear, if I have two real clusters in the dataset, and I fit 6 Gaussians, I expect to get 1-5 of them fall on top of one of the real clusters and the remaining 5-1 of them on top of the other cluster. This means looking at the distance matrix I expect to see quite a few (maybe 5) large distances [these are between-cluster distances] and lots of small distances [within-cluster distances].
here is a sample of the distance matrices I have, the dendrograms are just to help "see" the structure and have no additional information.
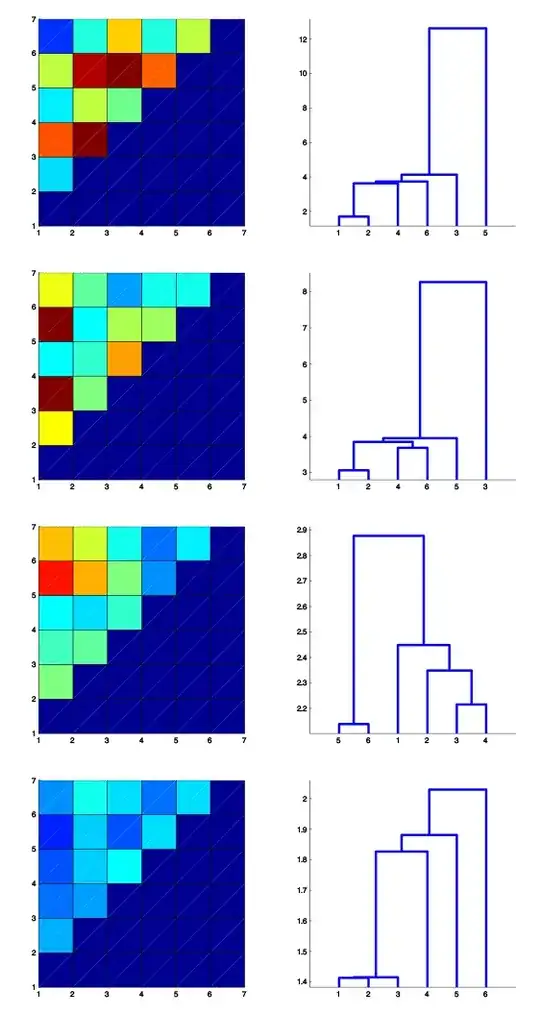
if anyone is interested in the raw(more raw) data, I can provide the data but if you don't have the domain specific knowledge (this is a spike sorting problem, in a neurophysiology context) it will not be easy for me to describe the data event in a few pages.
Any comments?