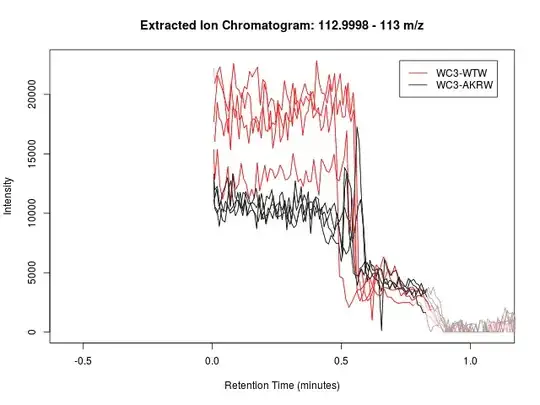
I have done an untargeted metabolomics study using LC-MS. There is one peak that matches to a biologically relevant compound but analyte eluted at the beginning of the LC run and the peak shape is far from ideal.
Do I assume the peak is an artifact, or can I say that there is a compound with that m/z in my sample? Ideally I would run the experiment again with standards, but I cannot do that due to time constraints. Someone else will follow up on the interesting compounds that I've identified and I wouldn't want to send them in the wrong direction.
Below is the peak in question, and following it is an example of a "good" peak from the same dataset.